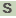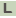>HMPREF9153_0359 riboflavin biosynthesis protein ribBA
MLNTIEEALAELRAGRPVLVADDADRENEGDAILAAELADPRWVGWMVRHTSGYLCAPMT
DEHADQLGLPLMWPANQDPLRTRYTVAVDAASGITTGIDAAQRARTARVLASATSTSADL
VRPGHVLPLRARAGGVLERRGHTEAAVDLARLAGLEPVGLIGELVGDDGMCLRTDSIAEL
ARTEDLAFVTIDQLAQWRLSHDPADTTAAPRVTALASAELPTRHGQFTMTGYRDNLTGAE
HVLLMSDKGIYADDGGPAWVRVHSECLTGDALGSLRCDCGDQLAAAQAHVNEHGGAIILL
RGHEGRATGLLNKIAAYRAQDGGLDTVDAQTHLGLPVDAREYGAAVAILADAGVSAVRLL
TNNPAKVDALREGGLEVTMSPLEIPARPEDERYLRTKRDRMGHLLALDHDMPEITTPDLN
HPEPARSTQGE
>HMPREF9153_0359 riboflavin biosynthesis protein ribBA
ATGCTGAACACCATCGAGGAGGCGCTGGCCGAGCTGCGCGCCGGACGTCCGGTGCTGGTT
GCCGACGACGCCGACCGGGAGAACGAGGGAGACGCCATCCTCGCCGCTGAGCTGGCTGAT
CCACGCTGGGTCGGATGGATGGTCCGCCACACTTCCGGGTACCTGTGCGCACCCATGACC
GACGAGCACGCTGACCAACTCGGTCTGCCCCTCATGTGGCCCGCCAACCAGGACCCGCTG
CGAACCCGCTACACCGTCGCCGTCGACGCGGCATCCGGGATAACGACCGGTATCGATGCC
GCCCAGCGTGCCCGCACCGCTCGAGTTCTCGCAAGCGCCACCTCGACCTCCGCTGACCTC
GTCCGTCCCGGCCACGTCCTGCCGCTACGGGCCCGCGCCGGGGGAGTGCTGGAGAGGCGC
GGTCACACCGAGGCCGCCGTCGACCTCGCCCGGCTGGCCGGCTTGGAACCCGTCGGTCTC
ATTGGTGAACTCGTGGGGGATGACGGTATGTGTCTGCGTACCGACTCCATAGCCGAGCTG
GCCCGCACCGAGGACCTGGCCTTCGTCACCATCGATCAACTGGCGCAGTGGCGGCTTTCC
CATGATCCGGCTGACACCACTGCGGCACCTCGCGTGACGGCCCTTGCCAGCGCGGAATTG
CCGACCCGGCACGGCCAATTCACGATGACCGGTTACCGGGACAACCTCACCGGGGCCGAA
CACGTCTTGCTCATGTCTGACAAGGGGATCTACGCCGACGACGGTGGCCCAGCCTGGGTG
AGGGTGCACTCGGAATGCCTCACCGGCGACGCCCTGGGATCCCTGCGCTGCGATTGCGGG
GACCAGCTGGCGGCTGCCCAGGCTCACGTCAACGAGCACGGCGGGGCCATCATCCTGCTT
CGTGGCCACGAGGGTCGAGCGACCGGTCTGCTCAACAAGATTGCGGCCTACCGGGCTCAG
GATGGTGGGCTCGACACCGTCGACGCCCAGACCCACCTCGGCCTGCCCGTCGACGCTCGC
GAGTACGGCGCGGCCGTGGCGATCCTGGCTGATGCCGGTGTTTCTGCCGTCCGCCTGCTC
ACGAACAACCCTGCCAAGGTGGATGCTCTGCGTGAAGGAGGACTTGAGGTGACGATGTCG
CCTCTCGAGATTCCGGCCCGTCCCGAGGACGAGCGGTACCTGCGTACGAAACGCGATCGG
ATGGGACATCTGCTGGCACTCGACCACGACATGCCCGAGATCACCACGCCCGACCTGAAC
CATCCTGAACCCGCCCGGAGCACCCAAGGAGAATGA