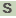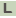>SACE_0755 cobalamin synthesis CobW-like protein
LPTEFGVGMHTEPSGAAGTDRRVPLVVVSGIHEEQVARAAEAVRARDAARTVVVQHDLRE
ISQGVVRRHVRHGDHEQLAVLELVHGCVSCTLREDFLPLLRRLCAAPDVERIVVRLDPAL
EPEAICWALQHVVVDGGTLDTDVAVEAVIAVVDRPSWLSDASGEDELAERGLAGSPDDER
TVAQVVVGQVEFADAVVLAGEAEDRWQEVRTEAVISRLTPTAPRGHLDGLDPADLLARVP
AEARRGRIDWPHGPLLRGQPSLESEAGVCVVLFEERRPFHPQRLHEVLDVLLDGVVRTRG
RAWVASQPGAALWLESAGGGLNVGHAGPWLAALDTAQWDEVSAERQVRASLNWDDYYGDR
MQELVVIAHEADPREITRALRSALLTDAEIAEGEAVWAGYEDPFGEWHTDPCPDMGAGTG
SEPATGSADAARDGGKQ
>SACE_0755 cobalamin synthesis CobW-like protein
TTGCCGACTGAGTTTGGGGTTGGGATGCACACCGAGCCGTCCGGTGCGGCGGGCACCGAC
CGGCGGGTCCCGCTGGTCGTGGTGTCCGGAATCCACGAGGAGCAGGTGGCGCGGGCAGCC
GAGGCGGTCCGCGCACGGGATGCCGCGCGCACCGTCGTCGTGCAGCACGACCTGCGCGAG
ATCTCCCAGGGCGTCGTCCGCAGGCACGTCCGACACGGCGACCACGAGCAGCTCGCGGTG
CTGGAACTGGTGCACGGCTGCGTGTCCTGCACGCTGCGCGAGGACTTCCTGCCGCTGCTG
CGCCGCCTGTGCGCCGCGCCCGACGTCGAGCGCATCGTGGTGCGGCTCGACCCGGCGCTG
GAGCCGGAGGCGATCTGCTGGGCGTTGCAGCACGTGGTGGTCGACGGCGGCACGCTCGAC
ACCGACGTCGCCGTCGAGGCCGTGATCGCGGTGGTCGACCGGCCCTCGTGGCTGTCCGAC
GCGTCGGGCGAGGACGAGCTGGCCGAGCGGGGCCTGGCGGGCAGCCCCGACGACGAGCGG
ACCGTCGCGCAGGTCGTGGTGGGACAGGTCGAGTTCGCCGACGCGGTCGTGCTGGCGGGC
GAGGCGGAGGACCGCTGGCAGGAGGTCCGCACCGAGGCGGTGATCAGCAGGCTCACCCCG
ACCGCGCCGCGCGGCCACCTCGACGGCCTCGACCCGGCGGACCTGCTCGCCCGGGTTCCC
GCGGAGGCGCGGCGGGGACGTATCGACTGGCCGCACGGTCCGCTGCTGCGCGGCCAGCCG
TCGCTGGAGTCCGAGGCCGGGGTCTGCGTGGTGCTGTTCGAGGAGCGCAGGCCGTTCCAC
CCGCAGCGGCTGCACGAGGTGCTGGACGTCCTGCTCGACGGCGTCGTGCGCACGCGCGGG
CGGGCGTGGGTGGCCAGCCAGCCCGGCGCGGCGCTCTGGCTGGAGTCGGCGGGCGGCGGC
CTCAACGTCGGCCACGCGGGACCTTGGCTGGCCGCGCTCGACACCGCGCAGTGGGACGAG
GTCTCGGCCGAGCGCCAGGTCAGAGCCTCGCTGAACTGGGACGACTACTACGGCGACCGG
ATGCAGGAACTGGTCGTCATCGCCCACGAGGCCGACCCGCGCGAGATCACCCGCGCACTG
CGCTCGGCACTGCTCACCGACGCCGAGATCGCCGAGGGCGAGGCGGTGTGGGCCGGCTAC
GAGGACCCGTTCGGCGAATGGCACACCGACCCGTGCCCCGACATGGGCGCGGGCACGGGC
TCGGAACCGGCCACCGGTTCCGCCGACGCCGCCCGCGACGGCGGCAAGCAGTGA